Simulation of frequency-dependent dielectric spectra¶
©️ Copyright 2025 @ Yao Wu
Author:
Yao Wu 📨
Date:2025-10-20
Lisence:This document is licensed under Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license.
1 Introduction¶
This tutorial is to give a guideline to calculate frequency-dependent dielectric spectra by using autocorrelation function and fourier transform of polarization fluctuation.
Dielectric constant is a quantitative measure of the electric polarizability of a dielectric material. It describes how much the electric field within the material is reduced relative to the vacuum when the material is placed in an external electric field.
The dielectric constant (or relative permittivity), denoted \(\varepsilon_{\mathrm{r}}\), is defined as the ratio of the electric displacement field D in the medium to the product of the vacuum permittivity \(\varepsilon_{\mathrm{0}}\) and the electric field E. Meanwhile, electric susceptibility is defined as the proportionality constant that relates an applied electric field E to the induced polarization density P, and is directly connected to the relative permittivity \(\varepsilon_{\mathrm{r}}\):
Note: all quantities below are expressed in SI units unless otherwise stated.
{Definition of Relative Permittivity} The relative dielectric constant is:
$$\varepsilon_{\mathrm{r}} = \frac{\mathbf{D}}{\varepsilon_{0}\mathbf{E}} = 1 + \chi $$ where \(\mathbf{D}\) is the electric displacement field; \(\mathbf{E}\) is the electric field; \(\varepsilon_{0}\) is the vacuum permittivity; \(\chi\) is the polarizability.
This formula shows that calculating the dielectric constant requires evaluating either the electric displacement under an electric field or the polarizability. Since calculations relying on electric displacement can only be valid in weak electric fields and fail to capture the frequency dependence of the dielectric constant, whereas polarizability can be obtained by statistical-mechanics methods, the polarizability route is more general.
Meanwhile,In statistical physics, the static dielectric constant of a dielectric can be calculated from equilibrium polarization fluctuations—this is the fluctuation–dissipation theorem (FDT) that links dielectric constant to polarization fluctuations: 1.The basic calculation method of dielectric constant
There are two ways to obtain the dielectric constant. The first relies on the polarization–electric-field relation and is valid only for weak fields. The second proceeds by computing the susceptibility. They are stated separately as follows:
where χ is the static electric susceptibility and \(\varepsilon_r\) the relative permittivity.
2.Fluctuation–dissipation theorem (static, zero field) For a cubic (isotropic) system at temperature T, susceptibility and polarization fluctuations are linked by
with $\langle \delta P^2 \rangle = \langle P^2 \rangle - \langle P \rangle^2 $ Inserting χ into \(\varepsilon_{\mathrm{r}}\) gives the dielectric-constant fluctuation formula:
3.Practical MD implementation The total dipole moment \(\mathbf{M} = \sum_i q_i \mathbf{r}_i\) is recorded during an NPT run. Since *\(P=M/V\)* and \(\langle \delta P^2 \rangle = \frac{\langle M^2 \rangle - \langle M \rangle^2}{V^2}\),
Averaging M over a sufficiently long equilibrium trajectory yields \(\varepsilon_{\mathrm{r}}\) without any external field.
Let us bring time dependence into the formula, the fluctuation of the total dipole moment M(t) be: \(\(\delta M(t) = M(t) - \langle M \rangle\)\)
Its time autocorrelation function (isotropic system, scalar form) is: \(\(C(t) = \langle \delta M(0) \cdot \delta M(t) \rangle\)\)
The frequency-dependent polarizability can be expressed as:
And \(F(C(t))\) is fourier transform \(F(t) = \int_{-\infty}^{\infty} f(t) e^{-i \omega t} dt\) .
Since time t and frequency \(\omega\) are reciprocally related, we will hereafter use frequency \(\omega\) instead. Subsequently, the real part of the polarizability \chi'(\omega) can be obtained via the Kramers-Kronig relations (Hilbert transform):
| Symbol | Meaning |
|---|---|
| χ′(ω) | The real part of the electric susceptibility at the target frequency ω. |
| χ″(ω′) | The imaginary part of the susceptibility, known as a function of the integration variable ω′. |
| ω | The frequency at which the real part is to be calculated. |
| ω′ | Integration variable running from 0 to ∞. |
| \(\mathcal{P}\) | Cauchy principal-value prescription, handling the pole at ω′ = ω. |
| 2/π | A constant prefactor fixed by the Kramers–Kronig relations. |
| Denominator (ω′² − ω²) | Weighting kernel that describes how susceptibility at other frequencies contributes to the real part at ω. |
Then, we can obtain the frequency information of dielectric constant as:
Below, we will take the simple BaTiO₃ (BTO) system as a test case and investigate its frequency-dependent dielectric spectrum.
2 Data preparation¶
MD simulation
The MD protocol should follow the same setup used for standard BaTiO₃ calculations, 10 * 10 * 10 supercell is used for simulation, Temperature T is set to 200 K.
As with most DeepMD simulations, you need to provide:
- Structure model as input file
- force-field model (DeepMD graph) as input file
- appropriate MD parametersls: time step, Tdamp(TAU_T), Pdamp(TAU_P), T, etc.
It is recommended to adopt the settings illustrated in the accompanying input.lammps template below. These parameters also can be adjusted as needed.
After this run you will obtain
log.200- LAMMPS log (thermo output)200trj.lammpstrj- corresponding trajectory file
The supercell volume and temperature written in log.200, as well as the polarization information stored in the .lammpstrj trajectory file, can be extracted automatically with the supplied parsing script from Li Denan's markdown.
Choose the total number of steps so that the entire trajectory covers the equilibration period plus the production time required for reliable polarization statistics. If the structure is not fully equilibrated (i.e. the average polarization has not stabilized), repeat the equilibration stage until the polarization fluctuates around a steady mean.
Adjust the thermo output interval to match the frequency range of interest, e.g. every 10, 20 or 50 steps.
- Writing every 10 steps (10fs) ⇒ f_max ≈ 50 THz
- Writing every 100 steps ⇒ f_max ≈ 5 THz
For higher-frequency response or finer high-frequency dielectric detail, use smaller output intervals (fewer steps between writes). In general: f_max = 1 /(2 × timestep)**
The total simulation length sets the lowest accessible frequency. With a 1 fs step and writes every 10 steps:
- 1 000 000 steps → 1 ns total → f_min ≈ 0.5 GHz
- 100 000 steps → 0.1 ns total → f_min ≈ 5 GHz
In general: f_min = 1 / (2 × total_time)
Input data preparation
Before start to calculate the autocorrelation function, make sure you have already finished the polarization calculation as a text file saved format with Px Py Pz columns. Px1 Py1 Pz1 means the start of first selected step of a MD trajectory. for example:
These polarization calculation method can be referred to in Liu yuan-jinsheng's and Li Denan's markdown file by using FerroDispCalc.(https://github.com/MoseyQAQ/FerroDispCalc/tree/cpp).
Additionally, the simulation temperature (T), the supercell volume (V) and timestep (step) ( between Px1 and Px2 is need to calculate dielectric spectra as input parameters.
3 Dielectric spectra calculation¶
Before computing any frequency-dependent dielectric response, we must emphasize that the entire trajectory must correspond to an equilibrium ensemble; otherwise the resulting spectra are physically meaningless.
We use the following script cal_dielectric_spectra.py with a CPU cluster to calculate the dielectric spectra.
If work well, the static dielectric constant will be print at screen and main output files will include the eps2.txt (dielectric spectra), Q (quality factor), tan(dielectric loss), nw (refractive index), kw(extinction function), Lostw(Energy loss). Or, please check your input data according to the bug report.
We also plot some results pictures of output files, the plotting script can be modified as needed.
Important notes:
- The input file must be corresponding with file_path parameter;
- Only T, V and step (Timestep)should be artificially input,
-
All the python modules should be correctly installed like cv2 and scienceplots;
-
nsmis the Gaussian-smoothing parameter for the data: the larger the value, the stronger the smoothing, but the more detail is lost. It is recommended to setnsm = (number of polarization rows) / 500 + 1. Additionally, it is recommended to setnsmto an odd number to ensure symmetric Gaussian smoothing and avoid potential off-by-one issues in the convolution kernel.
- Note: correct input parameters is needed to obtain accurate dielectric spectra !!!
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 | |
dielectric spectra results (eps2.dat && eps2.png):
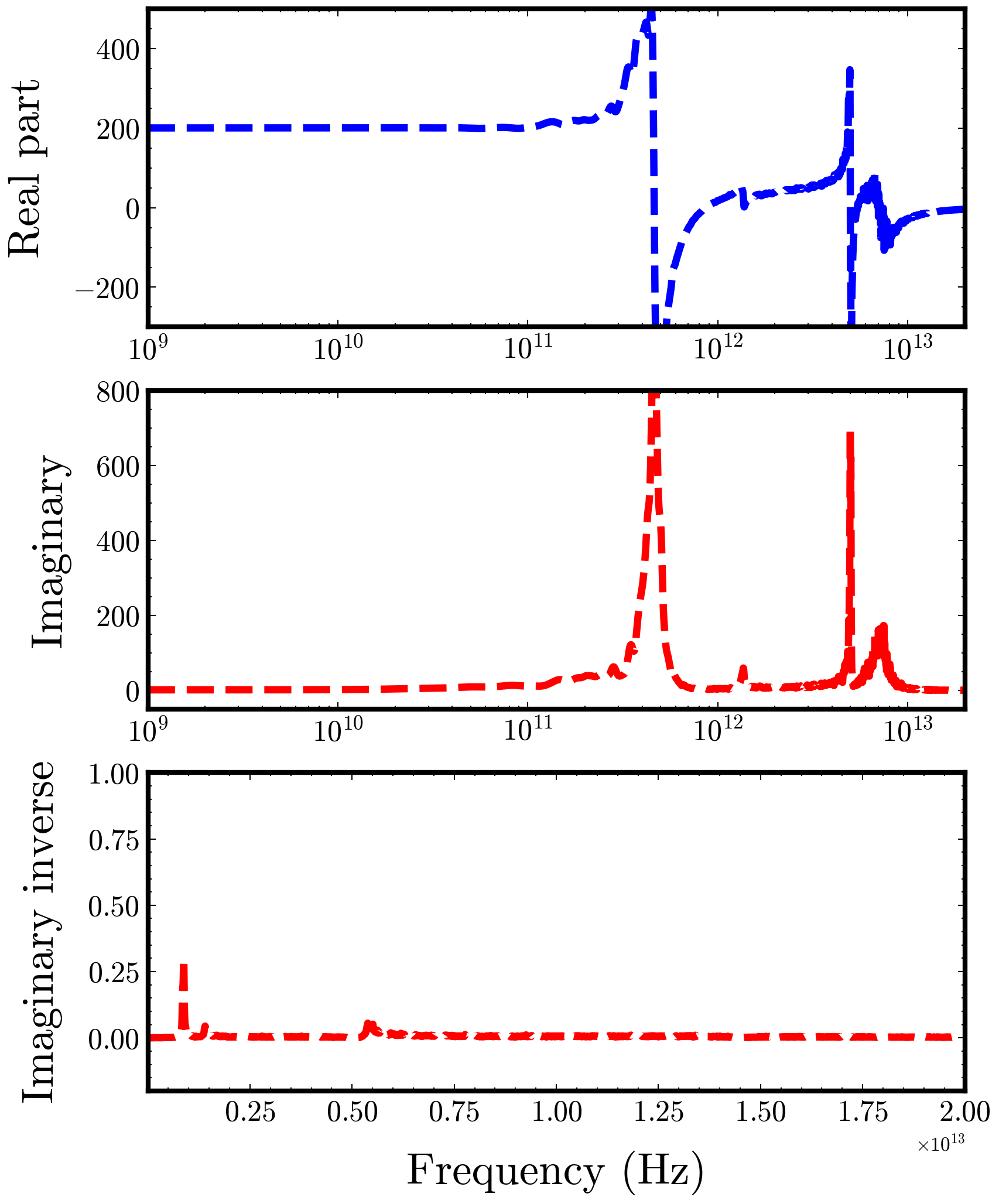
Note: Please double-check the paths and parameters in both the input files and the scripts before running. ****